New synthetic biology rule to modify antibiotics
Scientists from the Universities of Bristol, Birmingham and Leuven (Belgium) have decoded a key step in antibiotic production by bacteria, underpinning the development of new antibiotics to tackle the global threat from multiply resistant superbugs like MRSA and E. coli. Many important drugs, including antibiotics, are created by bacteria in a similar way (e.g. mupirocin, erythromycin, rifampicin). The discovery announced today brings us a step closer to controlling these biochemical pathways to produce novel molecules. For example, antibiotics might be modified to bypass the resistance mechanisms that superbugs use to survive in their presence.
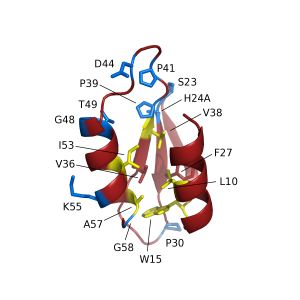
The process by which these complex chemicals are made is very much like a production line, with the molecule being extended piece by piece as it is passed along a huge, multi-functional protein. The team, comprising structural biologists, protein modellers, chemists and microbial geneticists, identified a code contained in the sequence of amino acids of the small carrier proteins that hold the growing antibiotic when it is being modified. Then using structural, computer and genetic analysis they worked out what parts of the carrier protein allows the modification machinery to identify and insert changes in precise areas of complex antibiotic structures. The code they identified consists of amino acids in the core of the protein as well as on its surface and applies to essentially all the biosynthetic factories of this type in any organism. The accompanying cartoon shows how the special carriers can dock with the modifying enzymes as an additional function the typical carriers do not have.
Tests showed that the carrier domains that have the correct code can be moved between systems but also that there are variants that allow two different modifications to be introduced at different positions in the same system. ‘This shows how we can direct this sort of modification when we are designing new synthetic pathways to make drugs against the increasingly problematic Enterobacteriaceae like Escherichia coli and Klebsiella pneumoniae,’ says University of Birmingham research lead Professor Chris Thomas.
Having the protein structure allowed the team make sense of the amino acid pattern identified. ‘These small carrier proteins appear to be quite simple and yet they seem to hold the key to creating new biosynthetic factories,’ adds University of Bristol research lead Professor Matt Crump.
Funded by the UK Biotechnology and Biological Sciences and Engineering and Physical Sciences Research Councils (BBSRC and EPSRC respectively) as well as the Darwin Trust of Edinburgh and the European Union, the researchers’ work paves the way for using synthetic biology create novel antibiotics that may help to solve the growing problem of bacterial infections that are resistant to essentially all antibiotics.
The research is published online in the journal Nature Chemical Biology.
For more information, please contact, Press Office, University of Birmingham, or Press Office, University of Bristol.
Notes to editors
“A conserved motif flags Acyl Carrier Proteins for b-branching in polyketide synthesis” byAnthony S Haines, Xu Dong, Zhongshu Song, Rohit Farmer, Christopher Williams, Joanne Hothersall, Eliza Płoskoń, Pakorn Wattana-amorn, Elton R. Stephens, Erika Yamada, Rachel Gurney, Yuiko Takebayashi, Joleen Masschelein, Russell J. Cox, Rob Lavigne, Christine L. Willis, Thomas J. Simpson, John Crosby, Peter J. Winn, Christopher M. Thomas, Matthew P. Crump. Nature Chemical Biology
For comments on the genetics aspect of this research please contact Professor Chris Thomas (c.m.thomas@bham.ac.uk). For comments on the structural biology aspects and chemical aspects of this research please contact Professor Matthew Crump (matt.crump@bristol.ac.uk).

Cartoon showing the green carrier domains that hold and pass on the antibiotic as it is being built. The carrier domains holding the antibiotic when it is modified have “pins” that fit the modifying enzyme, while the other ones are incompatible.