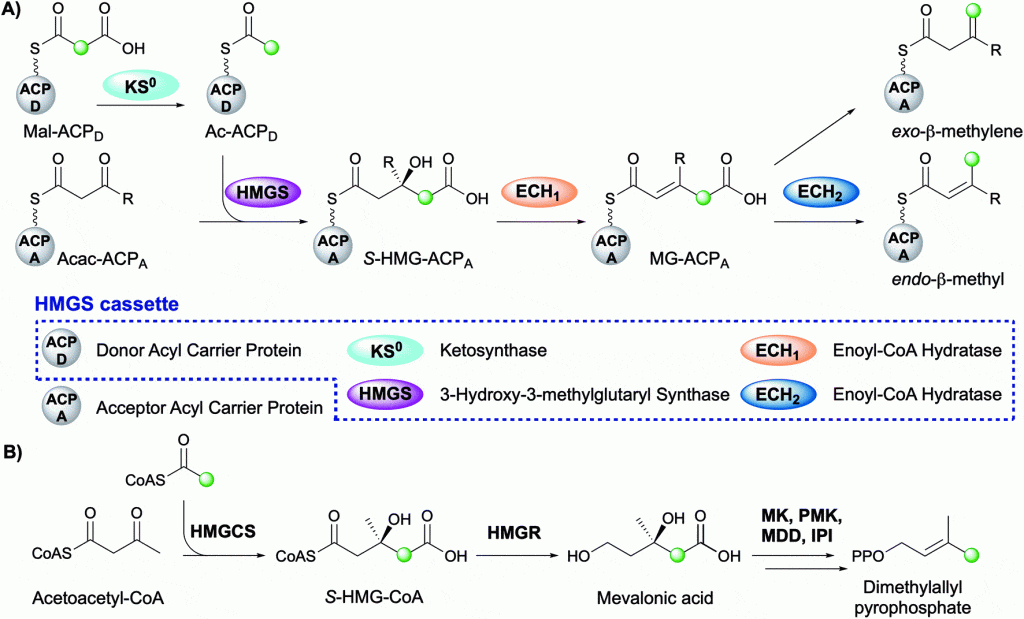
The Crump group is pleased to announce the publication of a recent review within Natural Product Reports. This review summarises the diversity of beta-branch incorporation within polyketide synthases and the mechanistic details of each catalytic step. The overall transformation is catalysed by a multi-protein 3-hydroxy-3-methylglutaryl synthase (HMGS) cassette and is reminiscent of the mevalonate pathway in terpene biosynthesis. The application of HMGS in data mining, additional beta-branching mechanisms and current knowledge of the role of beta-branches in this important class of biologically active natural products is also discussed.
Recent Paper: A Priming Cassette Generates Hydroxylated Acyl Starter units in Mupirocin and Thiomarinol Biosynthesis.
The Crump group is pleased to announce the publication of recent work in ACS Chemical Biology Journal. This work was completed as a collaboration between the Departments of Chemistry and Biochemistry at the University of Bristol, showing the need for frequent collaboration in the field of biosynthesis of natural products.
This work utilises a plethora of laboratory techniques to fully characterise the formation of a 3-carbon alcohol intermediate in the biosynthesis of the polyketide antibiotic mupirocin and homologous 4-carbon alcohol intermediate in the biosynthesis of thiomarinol.
This is exciting work and provides a solid contribution to the biosynthetic community, well done to all involed in the publication of this work.
A link to the paper can be found at the following link: https://doi.org/10.1021/acschembio.9b00969
Recent Paper: The Aminotriazole Antagonist Cmpd‐1 Stabilises a Distinct Inactive State of the Adenosine 2A Receptor
A very well done to everyone involved in this recent paper published in Angewandte Chemie. In this paper, 19F NMR has been used to study activation and inactivation of the A2A adenosine receptor. A compound with potential as an anti Parkinson’s drug was found to stabilise a novel inactive state of the A2A adensone receptor that is distinct from the standard inactive state observed in the literature.
The key finding of the paper is summarised in the below image.
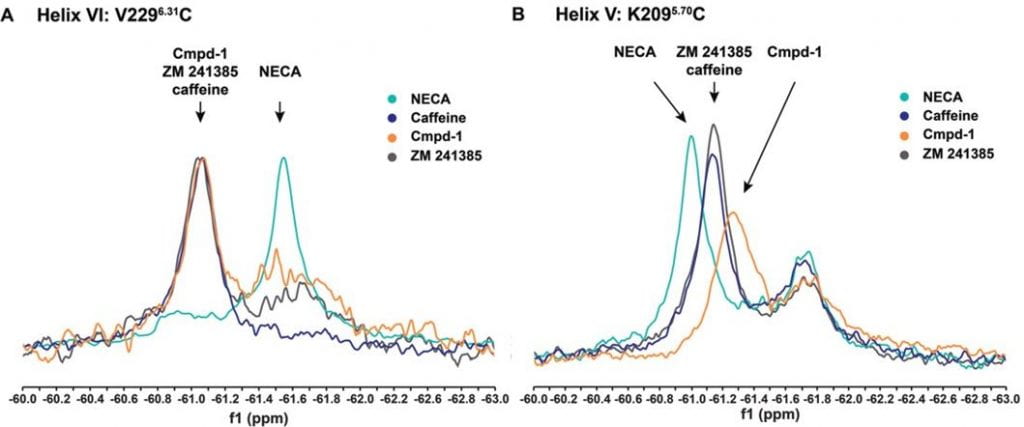 A) Normalised 19F NMR spectra of V2296.31C. A similar 19F chemical shift is observed when A2AR is exposed to three different antagonists, distinct from the agonist NECA B) 19F NMR spectra of K2095.70C. Cmpd‐1 causes the receptor to report a distinct state.
A) Normalised 19F NMR spectra of V2296.31C. A similar 19F chemical shift is observed when A2AR is exposed to three different antagonists, distinct from the agonist NECA B) 19F NMR spectra of K2095.70C. Cmpd‐1 causes the receptor to report a distinct state.
Paper can be found using the following link: https://doi.org/10.1002/anie.201902852
TrkA and NMR
After many years work our structural studies on the isolated TrkA domain that binds NGF has come to fruition with a new paper in J. Med. Chem. (See below). This study describes the use of an NMR construct of the second extracellular domain of the TrkA receptor to probe small ligand binding at the NGF binding site. This will be the basis of the development of further ligand scaffolds designed to agonise or antagonise the function of this receptor. We would be interested to hear from other groups working with this system who might wish to collaborate on future studies.
(Shoemark DK, Williams C, Fahey MS, Watson JJ, Tyler SJ, Scoltock SJ, Ellis RZ, Wickenden E, Burton AJ, Hemmings JL, Bailey CD, Dawbarn D, Jane DE, Willis CL, Sessions RB, Allen SJ and Crump MP* (2015) Design and Nuclear Magnetic Resonance (NMR) Structure Determination of the Second Extracellular Immunoglobulin Tyrosine Kinase A (TrkAIg2) Domain Construct for Binding Site Elucidation in Drug Discovery. J. Med. Chem. 58, 767-777. http://pubs.acs.org/doi/abs/10.1021/jm501307e)
New paper in Nature Chemical Biology
Local and macroscopic electrostatic interactions in single α-helices
Emily Baker,1 Gail Bartlett,1 Matt Crump,1 Richard Sessions,2,3 Noah Linden,4 Charl Faul1 and Dek Woolfson1,2,3
Nature Chemical Biology 11, 221 – 228 (2015)
DOI: 10.1038/nchembio.1739
http://www.nature.com/nchembio/journal/vaop/ncurrent/full/nchembio.1739.html
1School of Chemistry 2School of Biochemistry 3BrisSynBio 4School of Mathematics
From Dek: A multidisciplinary team of chemists, biochemists and mathematicians at Bristol have shed light on a controversial 40-year old theory of protein structure. The research was conducted by Emily Baker and Gail Bartlett working across the groups of Dek Woolfson, Charl Faul and Matt Crump, and in collaboration with Richard Sessions (Biochemistry) and Noah Linden (Mathematics). Briefly, many papers and textbooks describe how a macroscopic helix dipole can stabilise a-helices in proteins and influence protein folding, structure and function. Emily’s experiments, and analyses from Gail and Noah, show that the helix macrodipole is in fact vanishingly small compared with other electrostatic interactions that occur more locally in proteins.
A note from Matt: I should also point out that this study was only made possible through the NMR time allocated to us kindly by the Wellcome Trust funded HWB-NMR centre and access to the 900 MHz spectrometer that was essential for resolving the poorly dispersed proton (only!) spectra and taking me back to the good old days of two-dimensional sequential assignment!
Friday 19th December
Nearly the end of another year – its been a long one! But a high point of the last two weeks has been three new manuscripts accepted. The first is the emergence of our TrkA work in J. Med. Chem., (see pubs.acs.org/doi/abs/10.1021/jm501307e) where we have designed an NMR construct for use in drug discovery against this receptor. More to follow here I hope. There has also been a Nature Chemical Biology paper where my group has had a substantial involvement which will be published in the New Year. Finally a further paper in the long-standing successful collaboration with the Davis group which will be published in the New Year.
So lets see what next year brings – maybe Bruker will pull out of NMR as well? We have vowed not to buy any Agilent products in the future after their decision to do this and the hugely negative impact this has had on the NMR field and the impact on the huge research council investment now in legacy instrumentation all over the globe.
Studentship vacancy
Vacancies and Studentships.
We have a potential studentship working in the IGF2R domain 11 research project that would be aimed at engineering domain 11 for specific ligand binding. Recently we have used NMR, X-ray crystallography and yeast surface display assays to engineer an Insulin growth factor 2 (IGF2) super-antagonist based on domain 11 from the IGF2 receptor (IGF2R) (Crump & Hassan and co-workers 2012, Science 238, 1209-1213.) and the proposed PhD will build on this success by adapting domain 11 to a wider range of ligands. This studentship would be funded by the SWSBIO DTP (BBSRC) and would start for four years in October 2015. Please contact matt.crump@bristol.ac.uk for further details or see http://www.bristol.ac.uk/swdtp/projects_available/2015_projects_available/2015_wcb_bristol.html. Please note there will be an interview process for these studentships and final selection for a PhD will be based on this performance.
Supervisors: Prof. Matthew Crump (School of Chemistry, University of Bristol), Paul Race (School of Biochemistry, University of Bristol), Bass Hassan (Sir William Dunn School of Medicine, University of Oxford).
700 MHz micro-cryocoil II
The equipment is now due for deliver in February and we hope to be up and running shortly after this date. Please do not hesitate to contact me on (matt.crump@bristol.ac.uk) if you wish to access this facility. I will post specs in due course to give some idea of the sensitivity but in tests we have been able to detect traces of plasticiser as low as 30 nanograms in 1000 scans (about 20 minutes) at 600 MHz. We would therefore anticipate a 40% timesaving at 700 MHz 0r the ability to detect down to the 10 nanogram level in a few hours.
700 MHz Micro-cryocoil NMR
Events are moving ahead so we have now placed an order for the new 700 MHz Spectrometer equipped with a micro-coil cryoprobe (1.7mm). As part of BrisSynBio this will serve a number of projects but coming equipped with robotics for sample tube filling and a SampleJet for high-throughout, we encourage other interested parties to get in touch with me (matt.crump@bristol.ac.uk) to discuss possible applications for this instrument. We would like to explore in particular applications to high throughput structure determination of small molecules, metabolomics, high-throughput analyses of environmental samples as well as biological NMR applications.